The Power to Explore Biological Networks via Chemical Patterns
CARLSBAD is a database and knowledge inference system that integrates multiple bioactivity datasets in order to provide researchers with novel capabilities for the mining and exploration of available structure activity relationships throughout chemical biology space. The powerful cheminformatics and pattern recognition algorithms combined with network analysis methodology allow researchers using CARLSBAD to generate hypotheses involving the relationships between biological targets, chemical compounds and their common chemical patterns.
These capabilities have important implications for:
- Drug Repurposing
- Lead Compound Generation
- Side Effect Prediction
- Mode of Action Identification
- And More
Currently, CARLSBAD contains 755,329 compounds, 3613 protein targets, and 1,449,924 activities.
CARLSBAD is a database and knowledge inference system that integrates multiple bioactivity datasets in order to provide researchers with novel capabilities for the mining and exploration of available structure activity relationships (SAR) throughout chemical biology space. To learn more about CARLSBAD, refer to our publication:
The CARLSBAD platform consists of a database and web interface CarlsbadOne.
CarlsbadOne is a simplified web-based application streamlined to perform CARLSBAD’s most popular functions, lead discovery and off-target identification. CarlsbadOne utilizes the powerful structural algorithms, HierS and MCES, to help researches generate chemical pattern driven hypothesis.
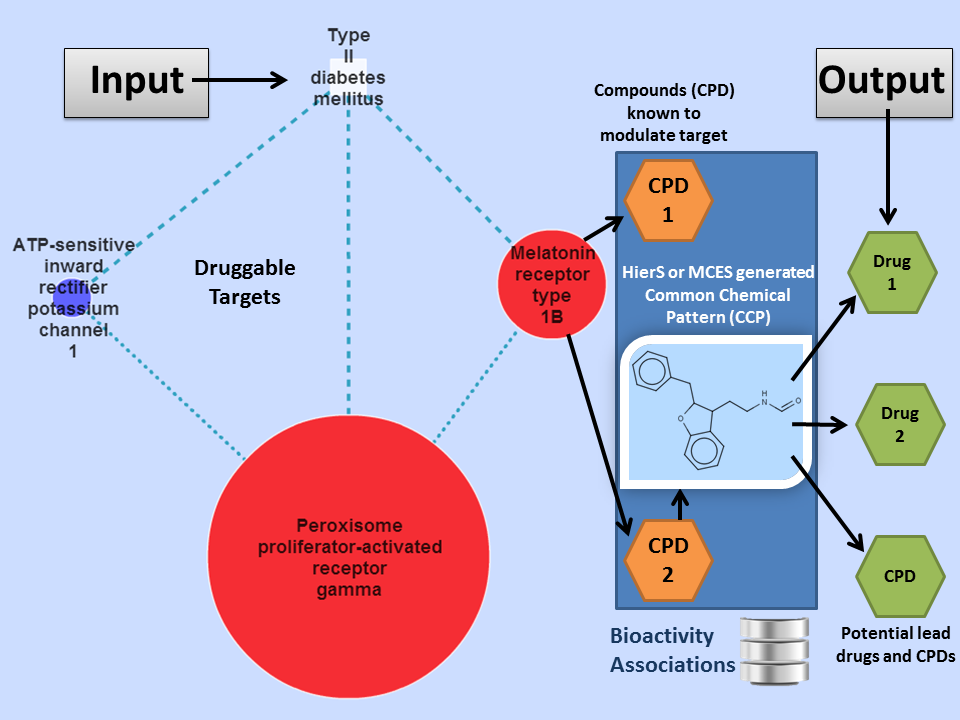
In one-click a user can query:
- Protein targets to determine potential lead compounds for that target.
- Drugs to determine potential off-targets for which that drug might be associated.
- Disease pathways to determine lead compounds to modulate druggable targets in a disease of interest.
Plus many useful features to facilitate analysis such as downloadable spreadsheets and knowledge graphs.
CarlsbadOne is a one-step, bioactivity data-mining, evidence-driven application for biophamaceutical knowledge discovery via cheminformatics, chemogenomics and systems pharmaco-informatics. CARLSBAD = Confederated Annotated Research Libraries for Small molecule BioActivity Data. This web app queries the CARLSBAD database, which consists of targets, small molecules, bioactivities, related data, and chemical patterns comprised of scaffolds and MCES clusters.
Query Logic
- Disease-query mode: A single disease is the query. Diseases are from KEGG. KEGG also reports linked genes,which reference proteins via NCBI_GI and Uniprot IDs, and thereby CARLSBAD targets.The one-click network includes those targets, active compounds, their CCPs, and neighbor compounds.
- Drug-query mode: A single drug is the query. All targets for which the drug is active are included in the retrieved subnet. In addition, targets are included for which the active compounds are associated with the drug query via chemical patterns.
- Target-query mode: A single protein target is the query. All active compounds are included in the retrieved subnet.In addition, compounds are included which are associated with the active compounds via chemical patterns.
Relationship to Query (R2Q) – Targets and compounds found each have a relationship to the query, and in Carlsbad these relationships are either:
- Empirical – related via empirical activity data.
- Hypothesis – related via common chemical pattern (CCP) and empirical data.
Relationships generally also vary in network distance. However, in CarlsbadOne, to be regarded as empirical, entities must be associated by only ONE link of each type (e.g. target→compound→target).
Reduced-graph representations – To facilitate effective visualization of large networks, reduced-graph representations are available:
- Targets-only: target-target edges represent shared compounds.Activity profile information is reflected via visual cues (e.g. target-node size).
- Targets-and-CCPs: compounds are hidden and represented via their CCPs. Target-target edges representing shared compounds included.
Output
- View summary of subnet.
- View a table of targets.
- View reduced-graph[s] subnet using CytoscapeWeb.
- View full subnet using CytoscapeWeb.
- Download XGMML file of reduced-graph[s] subnet.
- Download XGMML file of full subnet.
- Download a CSV file of subnet targets and associated data.
- Download a CSV file of subnet compounds and associated data.
The CarlsbadOne web app responds to the needs and feedback of users, by providing a unified portal, designed for usability and simplicity, with useful, relevant results for scientifically meaningful queries. Specifically, this means:
- One webapp UI for query and network viewing via CytoscapeWeb.
- One step, one topic query logic, with queries defined by one disease, drug, or target.
- One degree of separation via chemical pattern associations to novel target/compound hypotheses.
- One unified, coherent data resource.
- Step One for bioactivity knowledge discovery.
Development
CARLSBAD was developed by the University of New Mexico Translational Informatics Division, Department of Internal Medicine, and funded by NIH grant GM095952 (PI Tudor Oprea). CARLSBAD is offered free for academic and non-commercial use.
See also:
- GitHub repository: CARLSBAD
- Docker images: carlsbad_db, carlsbad_ui
- Carlsbad PostgreSql dump (available on request, for academic and non-commercial use).
